Data Science in Biomedicine
Research Group @ PLRI @ TU Braunschweig
Our group aims to elucidate the molecular mechanisms behind phenotypes and diseases. To that end, we develop integrative bioinformatics methods leveraging network analysis, machine learning, and statistics. We apply own and existing approaches in close collaboration with biologists and physicians to derive insights from multi-omics data.

News
Projects

PRETTY
The PRETTY (Personalised Prediction of Transplant Toxicity) project aims to develop a personalized prediction model to assess the risk of serious kidney damage following stem cell transplants in leukaemia patients. Its goal is to reduce the risk factors and thereby increase the lifespan and quality of life of those affected.
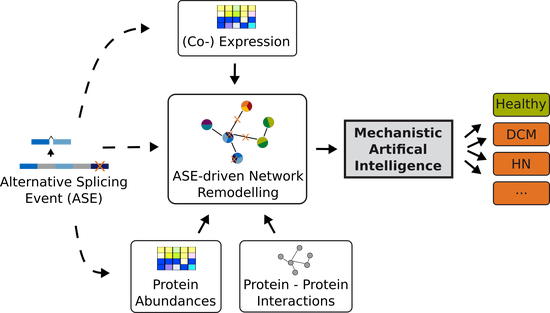
Sys_CARE
Systems Medicine Investigation of Alternative Splicing in Cardiac and Renal Diseases.

REPO-TRIAL
The EU H2020 project REPO-TRIAL aims at developing an in silico approach to optimise the efficacy and precision of drug repurposing trials. To this end we integrate heterogeneous data into a comprehensive interactome of disease-drug-gene interactions (a new diseasome) and develop graph-based machine learning approaches to investigate this highly complex data.

FeatureCloud
The EU H2020 project FeatureCloud aims at developing methods for privacy-preserving, federated machine learning.
Network-Based Epistasis Detection
We tackle the challenge of higher-order epistasis detection using biological networks to narrow the search space and GPU computing to improve the efficiency. Phenotype-specific epistasis-modules extracted from larger networks will help to better understand the underlying biological mechanisms of different phenotypes.
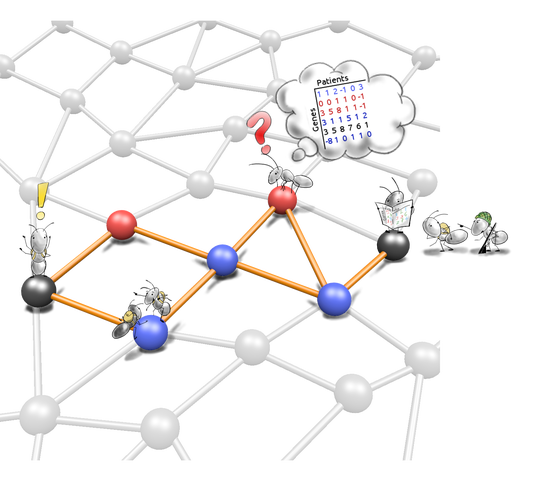
De novo network enrichment
We develop tools that leverage information from molecular interaction networks in understanding molecular profiling data. De novo network enrichment tools extract subnetworks that mechanistically explain a phenotype of interest, e.g. a disease.
Software & Resources

Epistasis Disease Atlas
The EDA is a comprehensive database focused on epistatic interactions, analyzing how groups of genes work together to cause complex diseases. The atlas currently covers Alzheimer’s, rheumatoid arthritis, bipolar disorder, coronary heart disease, inflammatory bowel disease, type 1 and type 2 diabetes, and hypertension. Data is accessible through a user-friendly R-Shiny app for searching, visualization, and analysis.
ASimulatoR
ASimulatoR is an R package for the simulation of RNA-seq reads with alternative splicing events. It is freely available on GitHub.

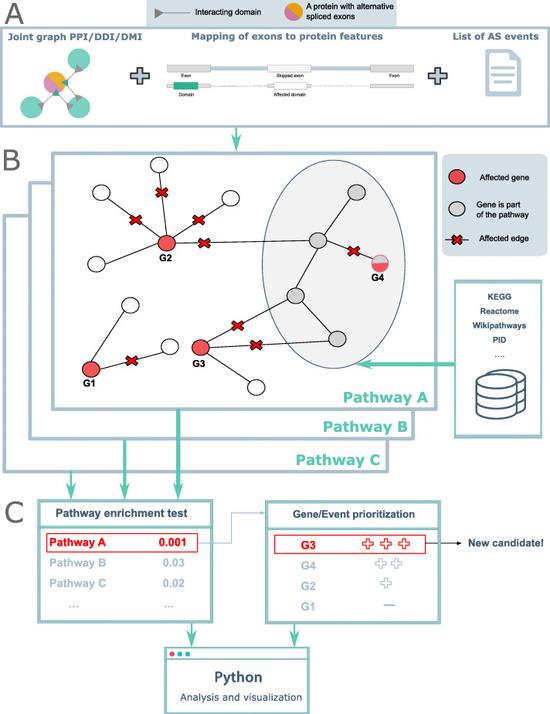
DIGGER
Domain Interaction Graph Guided ExploreR (DIGGER) integrates protein-protein interactions and domain-domain interactions into a joint graph and maps interacting residues to exons. DIGGER allows the users to query exons or isoforms individually or as a set to visually explore their interactions.
CoVex
To address the pandemic of the Coronavirus Disease-2019 (COVID-19), drug repurposing can be a helpful approach since it offers the possibility to find alternative fields of application for already approved drugs. CoVex is the first network and systems medicine online data analysis platform that integrates virus-human interaction data for SARS-CoV-2 and SARS-CoV. It is available as interactive webtool. More information and current updates can be found at the CoVex blog at the Chair of Experimental Bioinformatics website.

Scellnetor
Scellnetor is a novel scRNA-seq clustering tool. It allows the analysis of pseudo time-courses in single-cell sequencing data via a network-constrained clustering algorithm. Scellnetor is available as interactive online application at the Scellnetor website.
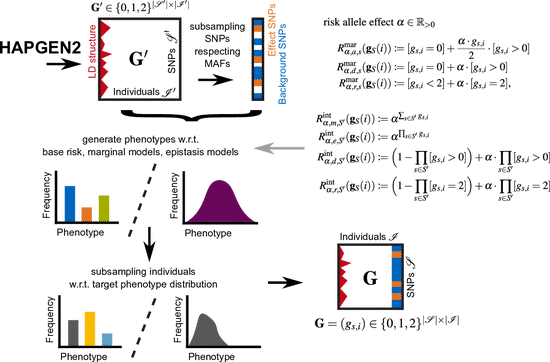
EpiGEN
EpiGEN is a Python pipeline for simulating epistasis data. It supports epistasis models of arbitrary size, which can be specified either extensionally or via parametrized risk models. Moreover, the user can specify the minor allele frequencies (MAFs) of both noise and disease SNPs, and provide a bias target distribution for the generated phenotypes to simulate observation bias. EpiGEN is freely available as python 3 package on GitHub.
Fastlogranktest
Fastlogranktest is a software package providing wicked-fast implementations of the logrank test in C++, R, and Python.
BiCoN
BiCoN is a powerful new systems medicine tool to stratify patients while elucidating the responsible disease mechanisms. BiCoN is a network-constrained biclustering approach which restricts biclusters to functionally related genes connected in molecular interaction networks and maximizes the expression difference between two subgroups of patients. A package for network-constrained biclustering of patients and multi-omics data can also be used. Download and installation instructions can be found here.